
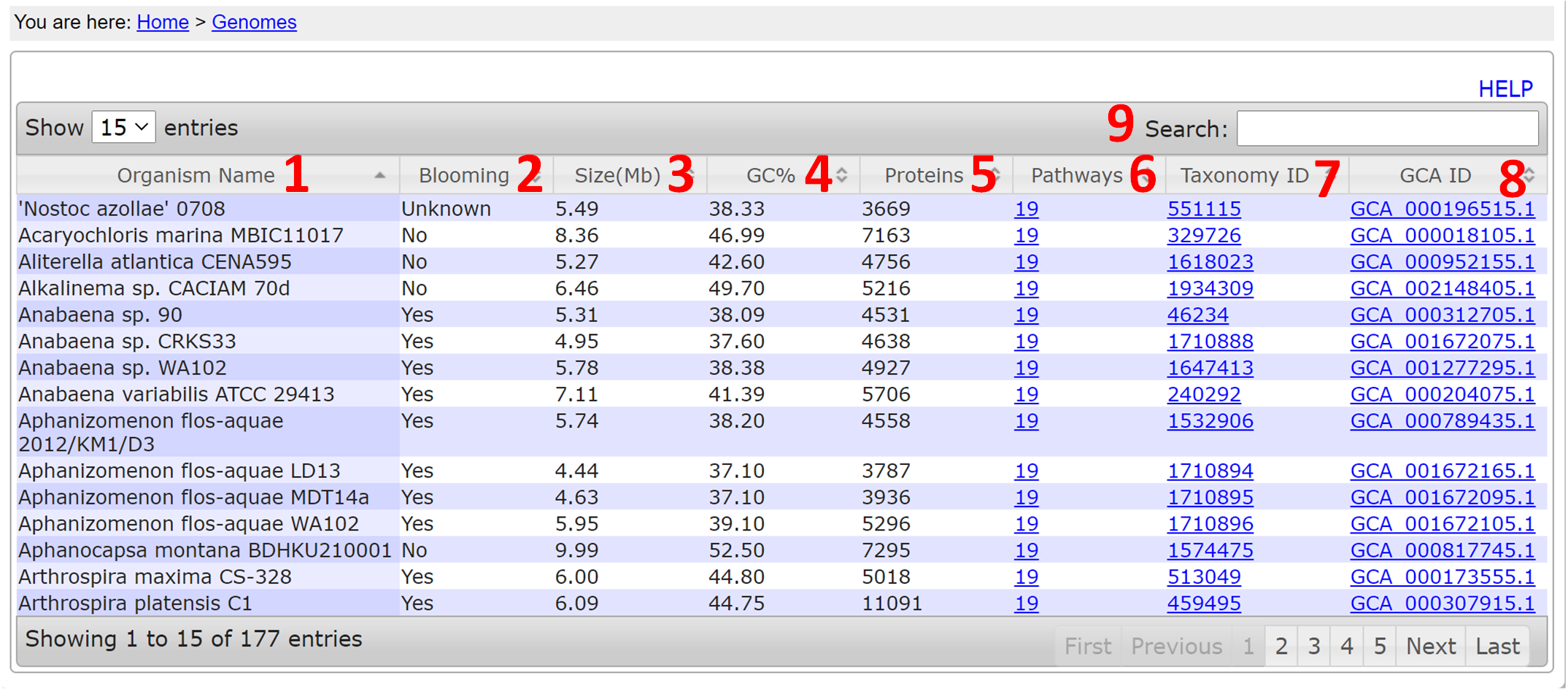
- Organism Name: The name of the species
- Blooming: Whether the species forms blooms
- Size(Mb): Genome size
- GC%: GC content
- Proteins: The number of proteins in the species
- Pathways: The number of pathways in the species
- Taxonomy ID: Taxonomy ID from NCBI
- GCA ID: Genome assembly number
- Search: typing in any letters/words can search any columns of the table.
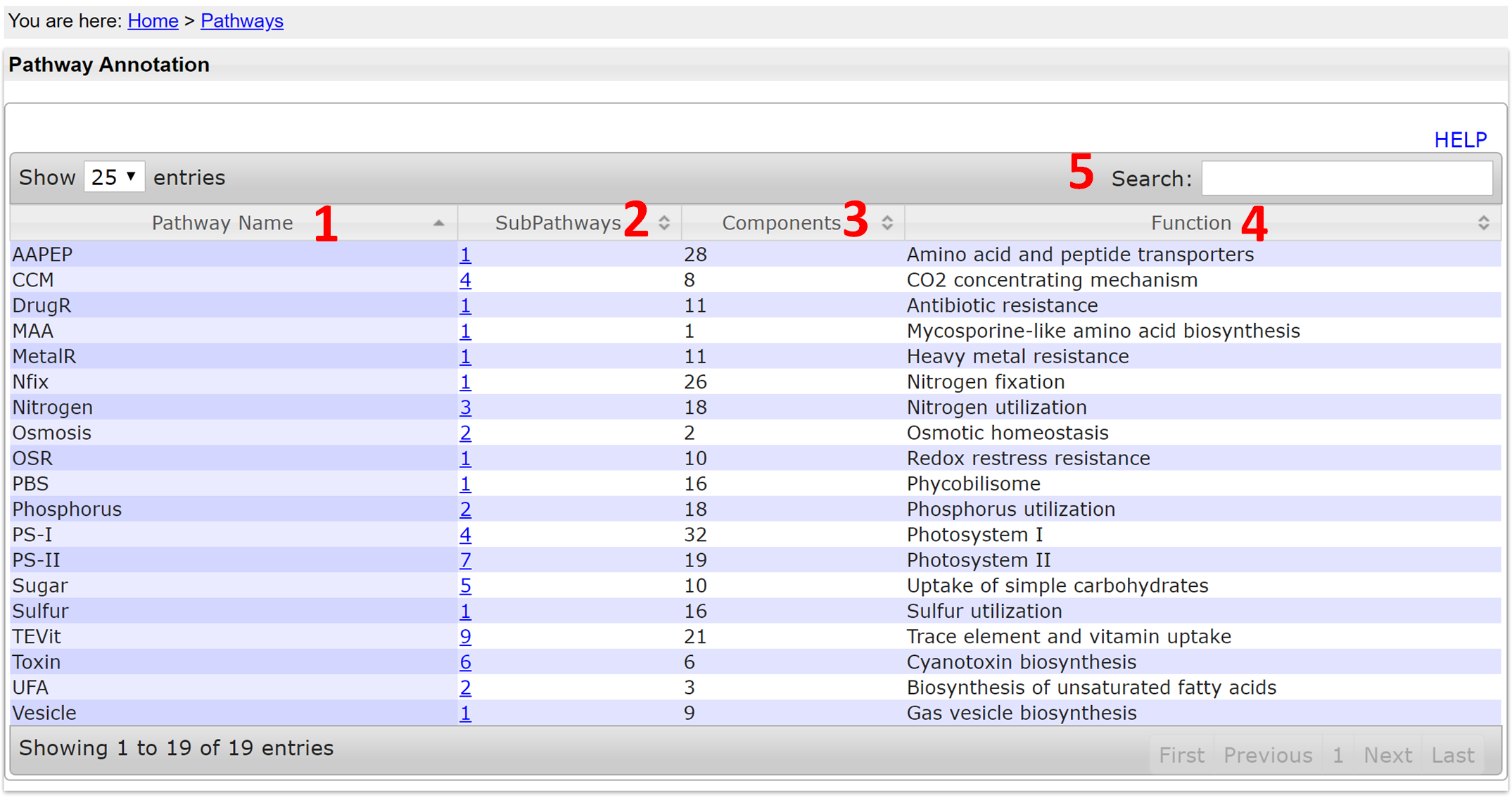
- Pathway Name: The name of the pathway
- SubPathways: The subpathway of a pathway
- Components: The proteins (multi-/single-protein complexes) in a pathway
- Function: The function of the pathway
- Search: typing in any letters/words can search any columns of the table.
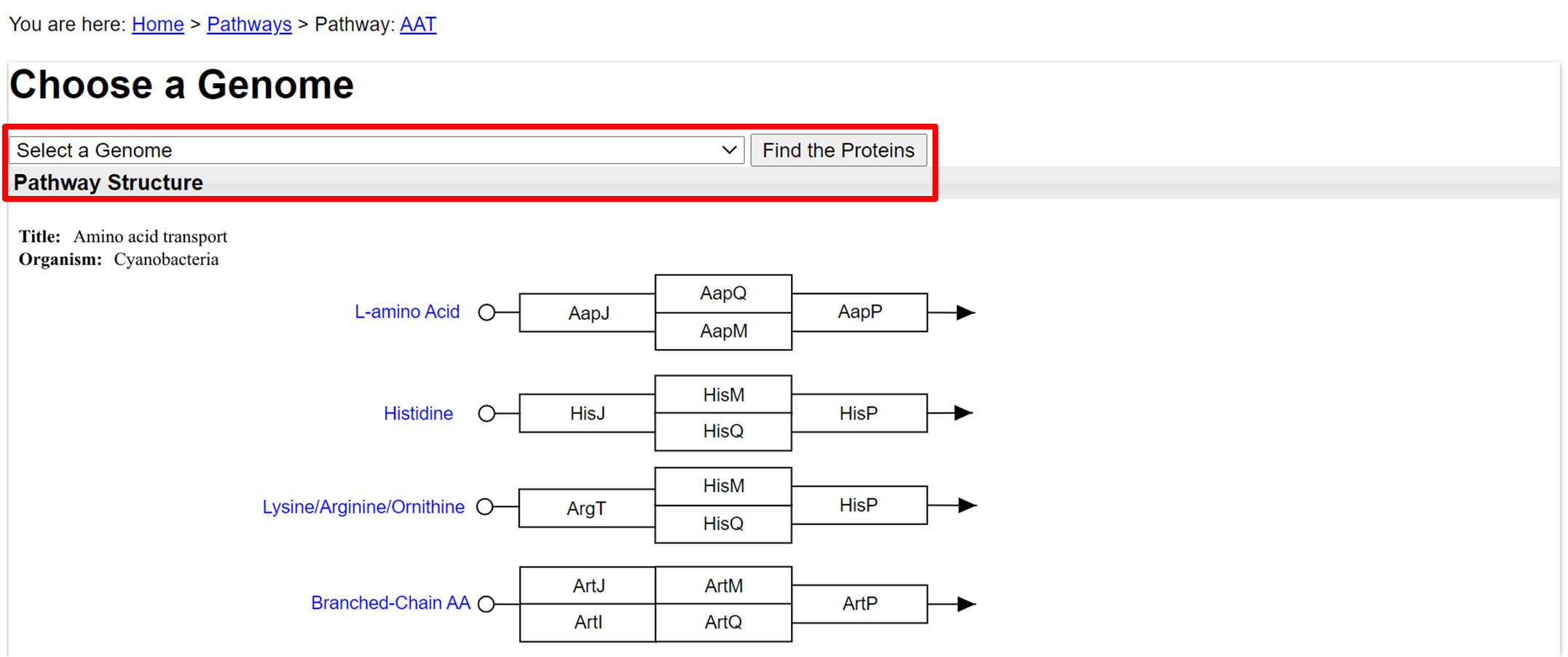
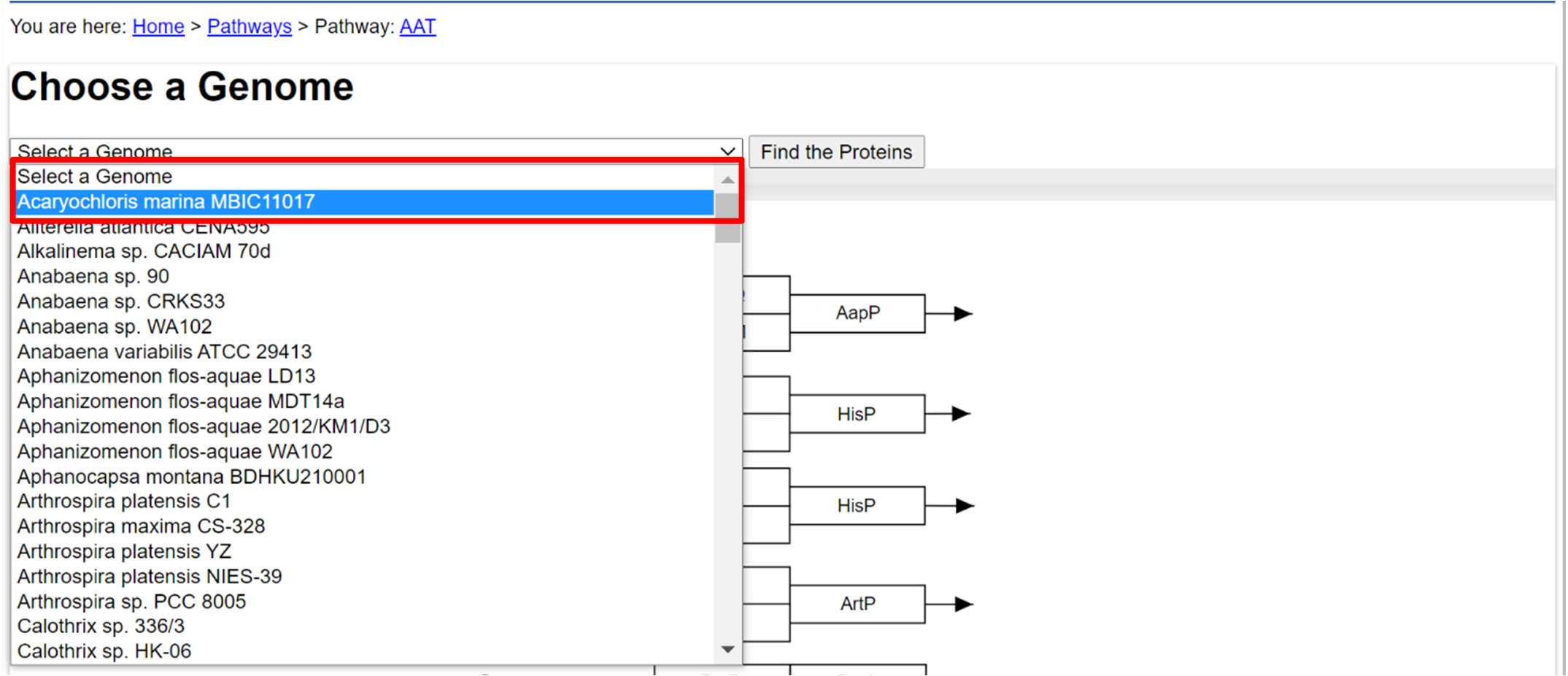
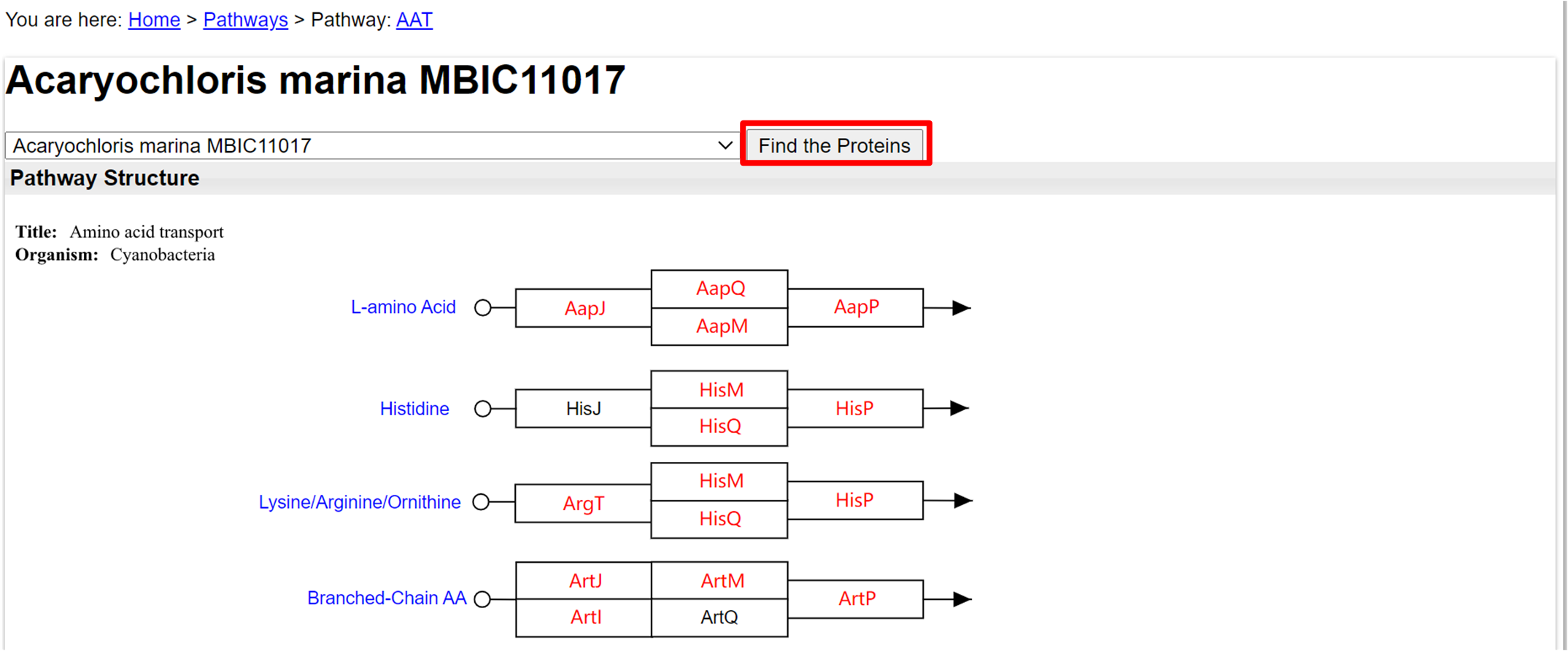
- In a selected protein pathway, at the top bar choose a genome.
- Click on the genome that you want to blast with the proteins.
- Click "Find the Proteins" to have the proteins found in that genome to be sequence highlighted.

- The system will send a Email to the submitted address when the prediction are finished;
- Input some sequences of proteins as FASTA format;
- Upload a fasta file including many sequences of proteins.

- The raw CyanoPATH output; right click to download;
- The tabular output of CyanoPATH prediction;
- The parsed CyanoPATH result:
(1) Query name:The name of input proteins;
(2) Pathway:The prediction results of Pathway;
(3) SubPathway:The prediction results of SubPathway;
(4) Component:The prediction results of Component;
(5) Evalue:E-value of Blast;
- Search: typing in any letters/words can search any columns of the table.







